How Vector Databases Search: A Practical Guide to IVF, HNSW, PQ & ScaNN

Introduction
Modern applications-from image similarity to RAG systems-run on fast vector search. When you embed text, audio, or images into high‑dimensional vectors, finding the closest matches becomes a problem of approximate nearest neighbor (ANN) search. This is where vector databases like FAISS, Milvus, Pinecone, Weaviate, and Elasticsearch step in.
But behind the scenes, these systems use very different indexing strategies. Some are clustering‑based (IVF), some graph‑based (HNSW), some compress aggressively (PQ), and some are hyper‑optimized via learned heuristics (ScaNN).
This post breaks down how these indexes work, why each exists, and how to choose the right one-without drowning in academic theory.
We'll use intuitive examples, simple diagrams, and real‑world constraints from Hoomanely’s own RAG + retrieval systems.
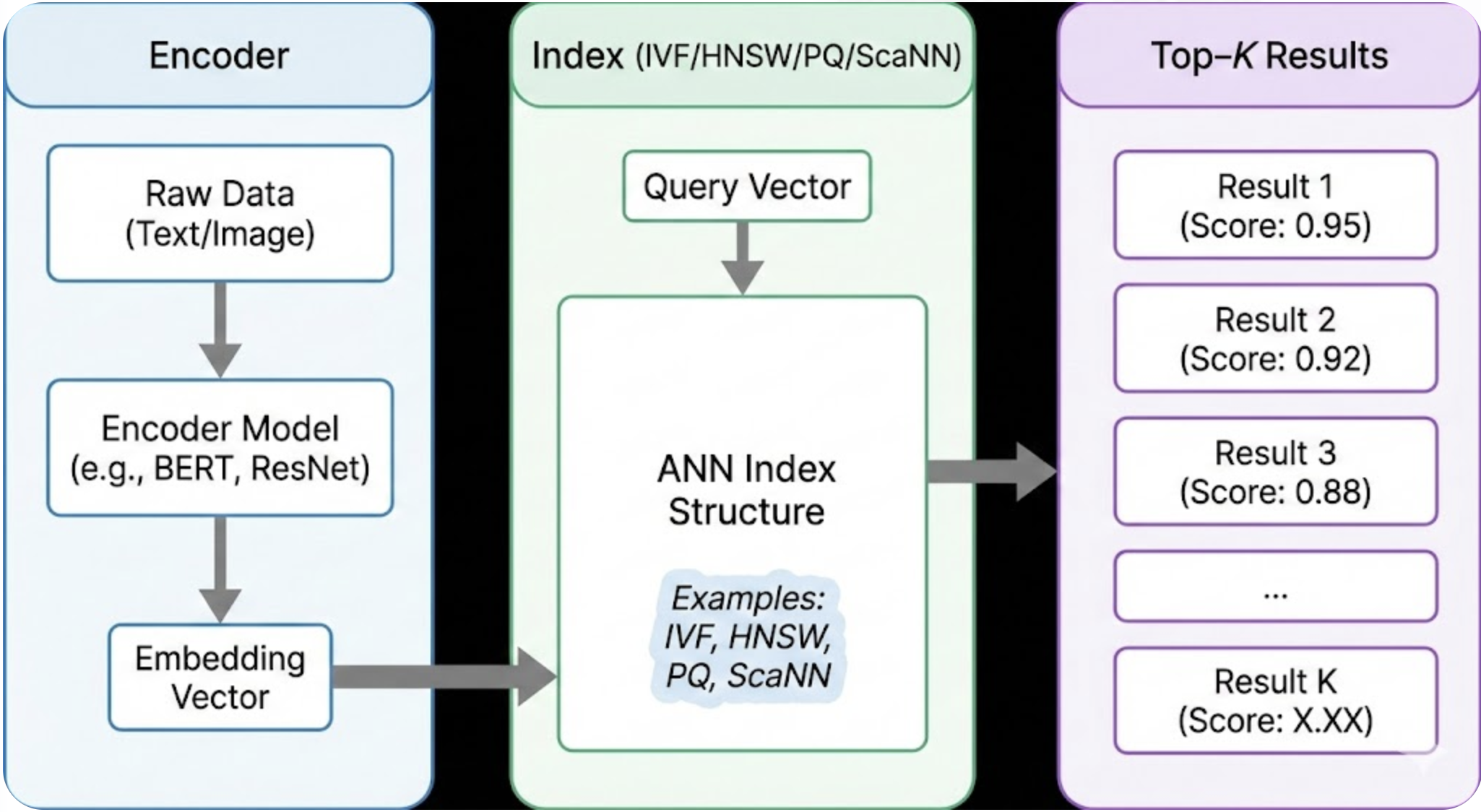
Why Vector Indexes Matter
A naive vector search scans every embedding-O(N) complexity. For a million vectors, that’s trivial; for a hundred million, your latency explodes.
ANN indexes accelerate search by:
- Cutting the search space (IVF)
- Navigating only promising regions (HNSW)
- Compressing vectors to fit in RAM (PQ)
- Reducing compute via learned pruning (ScaNN)
In systems like Hoomanely, where we handle thousands of pages per query across hundreds of veterinary PDFs, low-latency retrieval is core to a smooth user experience.
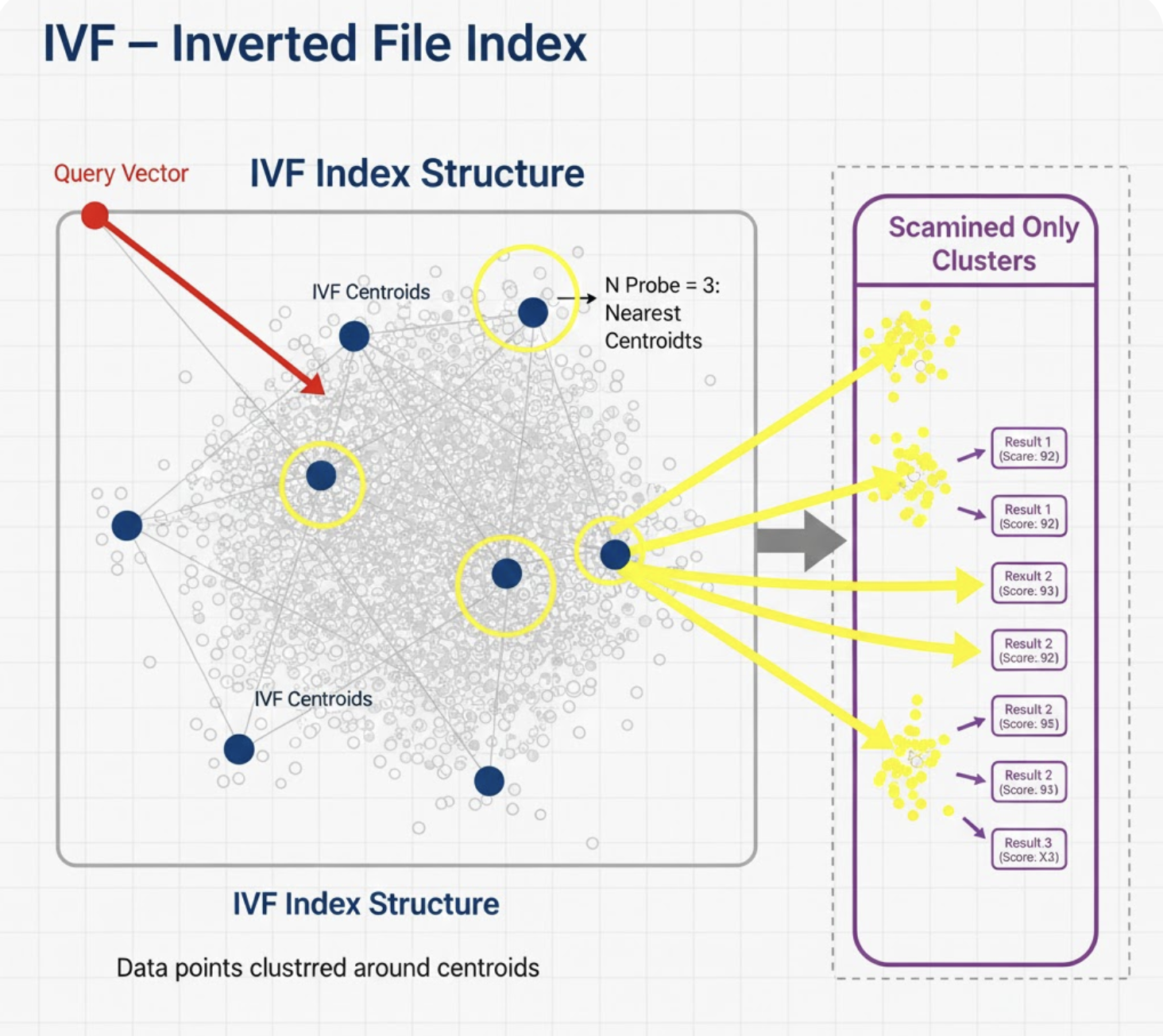
1. IVF - Inverted File Index
Concept
IVF partitions the vector space using clustering (usually k-means). Instead of searching all vectors, you search only a few clusters.
Think of it like a library:
- First, shelves (clusters) are numbered.
- When you search, you only walk to a handful of shelves.
How It Works
- Run k-means to get nlist centroids.
- Assign every vector to the nearest centroid.
- At query time:
- Assign query to nearest centroids.
- Search vectors only in those partitions.
Mini Example
- You create 1,024 clusters.
- For each search, you probe 8.
- That’s a 128× reduction in search cost.
Pros
- Very memory‑efficient.
- Easy to scale.
- Works great with PQ (IVF‑PQ).
Cons
- Quality depends heavily on clustering.
- Not ideal for unstructured distributions.
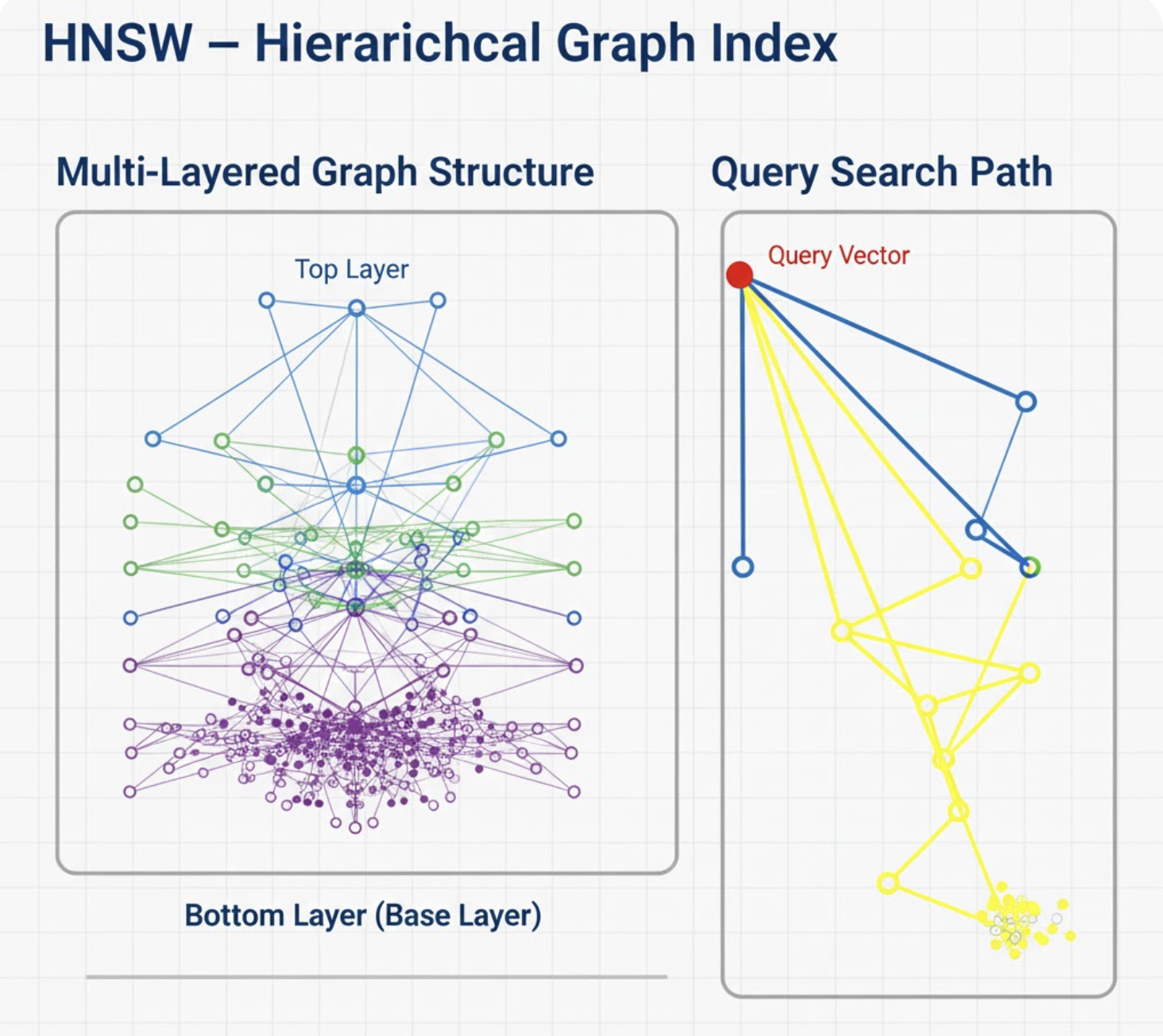
2. HNSW - Hierarchical Graph Index
Concept
HNSW builds a multi‑level navigable small‑world graph. Each node links to its nearest neighbors. Higher levels provide long‑range jumps; lower levels provide fine precision.
Imagine Google Maps but with flyover highways (top layers) + local roads (bottom layers).
How It Works
- Build multiple layers; each upper layer has fewer nodes.
- Insert each vector with random layer height.
- During search:
- Start at top layer → move greedily toward the query.
- Drop layers until you reach the ground layer.
- Explore neighbors and return top‑K.
Pros
- Highest recall at low latency.
- Great for real‑time updates (insert/delete).
- No expensive clustering.
Cons
- Higher memory footprint.
- Slow to build for large datasets.
Used by: Pinecone, Milvus, Weaviate, FAISS-HNSW.
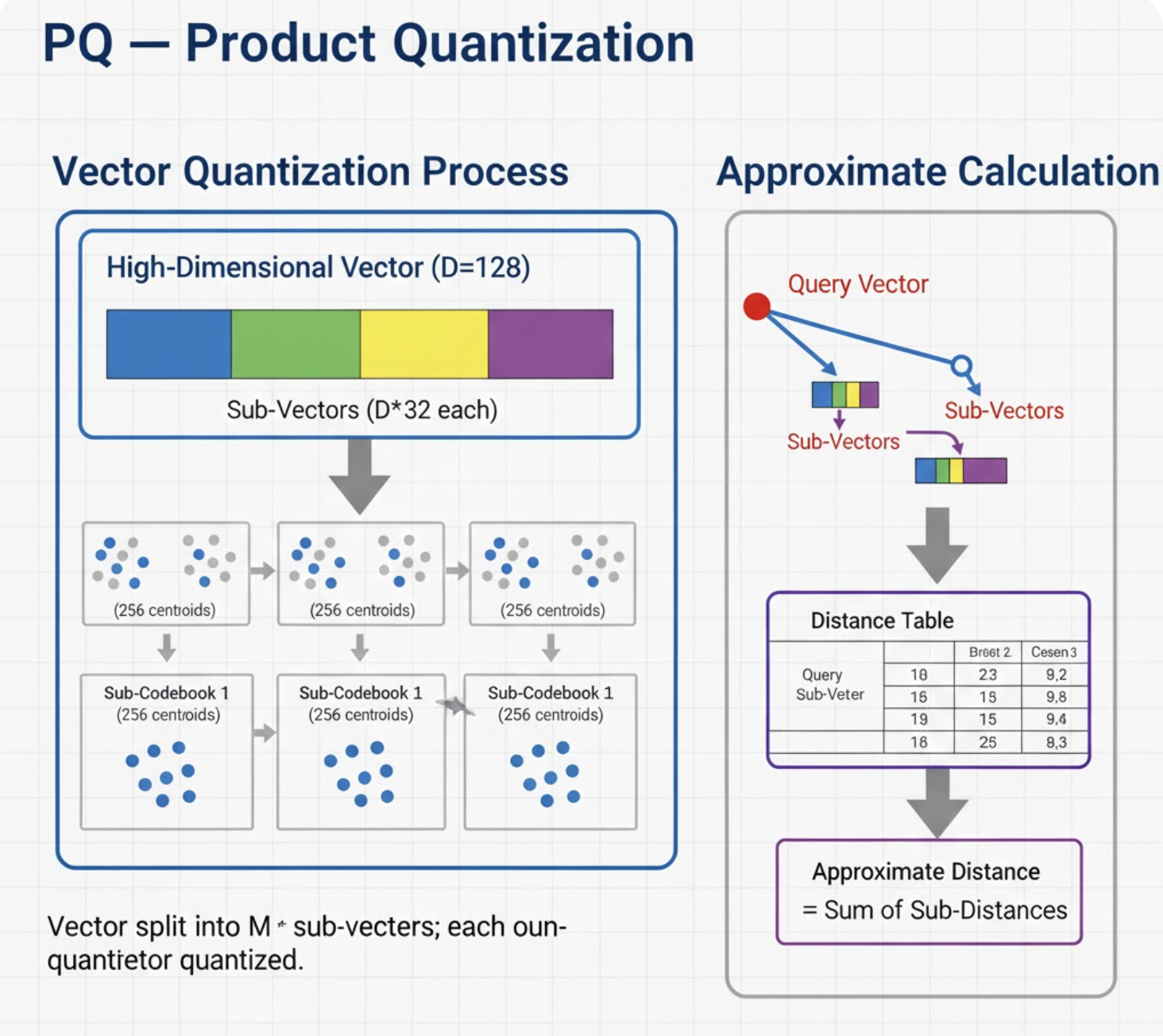
3. PQ - Product Quantization
Concept
PQ compresses each vector into small discrete codes using quantization. You trade accuracy for significant memory savings.
Think of splitting a 768‑dim vector into 16 chunks of 48 dims each, then encoding each chunk using a small lookup table.
How It Works
- Split each vector into m sub‑vectors.
- Cluster each subspace separately.
- Store only the cluster IDs.
- During search:
- Precompute distances to all codebooks.
- Estimate distances very fast.
Pros
- Compresses vectors by 8×–32×.
- Allows massive datasets to fit in RAM.
- Works well with IVF (IVF‑PQ).
Cons
- Lower recall compared to HNSW.
- Codebook training matters a lot.
Used by: FAISS‑PQ, Milvus IVF‑PQ.
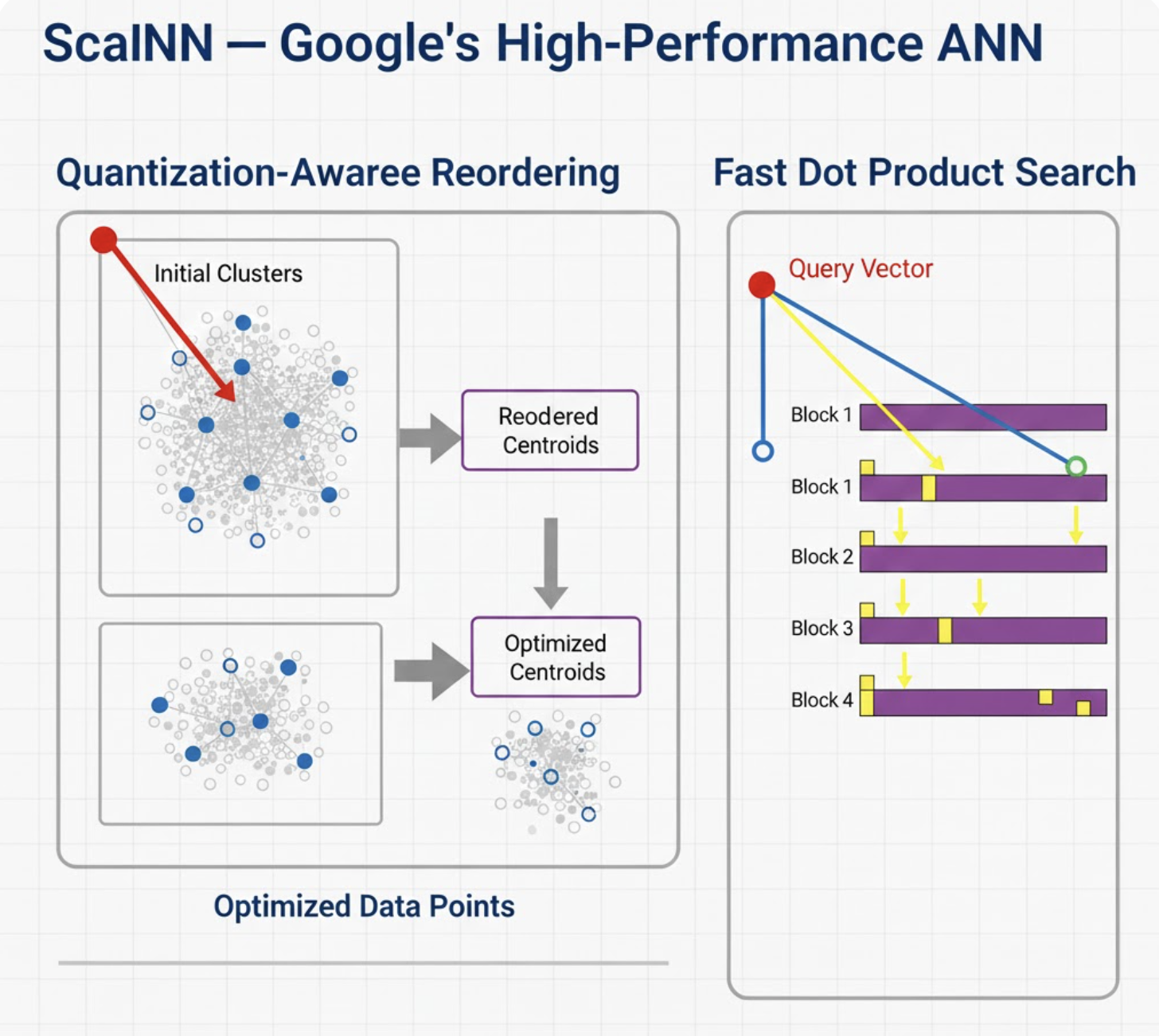
4. ScaNN - Google’s High‑Performance ANN
Concept
ScaNN (pronounced “scan”) mixes tree partitioning, anisotropic quantization, and learned pruning. Its strength lies in balancing speed and accuracy without excessive memory use.
How It Works
ScaNN typically uses:
- Partitioning: tree‑like IVF to narrow search.
- Score quantization: faster distance computations.
- Residual reordering: rescoring top candidates exactly.
This makes it lightning‑fast on TPUs/CPUs.
Pros
- High recall with moderate memory.
- Great for text embeddings.
- Strong performance in batch mode.
Cons
- Harder to tune.
- Not as common in production as HNSW.
Used in: Google Search, Vertex Matching Engine.
Comparing All Four
| Index | Best For | Memory | Build Time | Recall | Typical Use |
|---|---|---|---|---|---|
| IVF | Large static datasets | Medium | Medium | Medium | RAG systems, batched search |
| HNSW | Real‑time, high‑recall search | High | Slow | High | Recommendations, semantic search |
| PQ | Huge datasets with tight RAM | Very Low | Medium | Low‑Medium | Image search, dedup, RAG archives |
| ScaNN | High‑speed text search | Medium | Fast | High | Web‑scale retrieval |
Image Placeholder: "Spider chart comparing HNSW, IVF, PQ, and ScaNN across Recall, Speed, Memory, Ease."
How We Apply These at Hoomanely
At Hoomanely, our mission is to empower pet parents with real‑time, AI‑powered insights. A core part of this is our veterinary RAG pipeline, which retrieves the right pages from thousands of medical documents.
We’ve tested IVF, HNSW, and PQ combinations extensively. In internal benchmarks:
- HNSW provides the strongest recall, especially for long clinical queries.
- IVF‑PQ gives the best memory footprint, ideal for large document sets.
- Hybrid setups (IVF‑HNSW) give fast retrieval on CPU‑only environments.
Choosing the right index helps us deliver faster answers and more accurate guidance for pet wellbeing.
Takeaways
- IVF: great first choice for scalable, static datasets.
- HNSW: best all‑rounder for high recall + dynamic inserts.
- PQ: use when memory is tight or datasets are huge.
- ScaNN: ideal for high‑speed text retrieval, especially in cloud environments.
The “best” index depends on your constraints: memory, latency, recall needs, and whether vectors change frequently.





